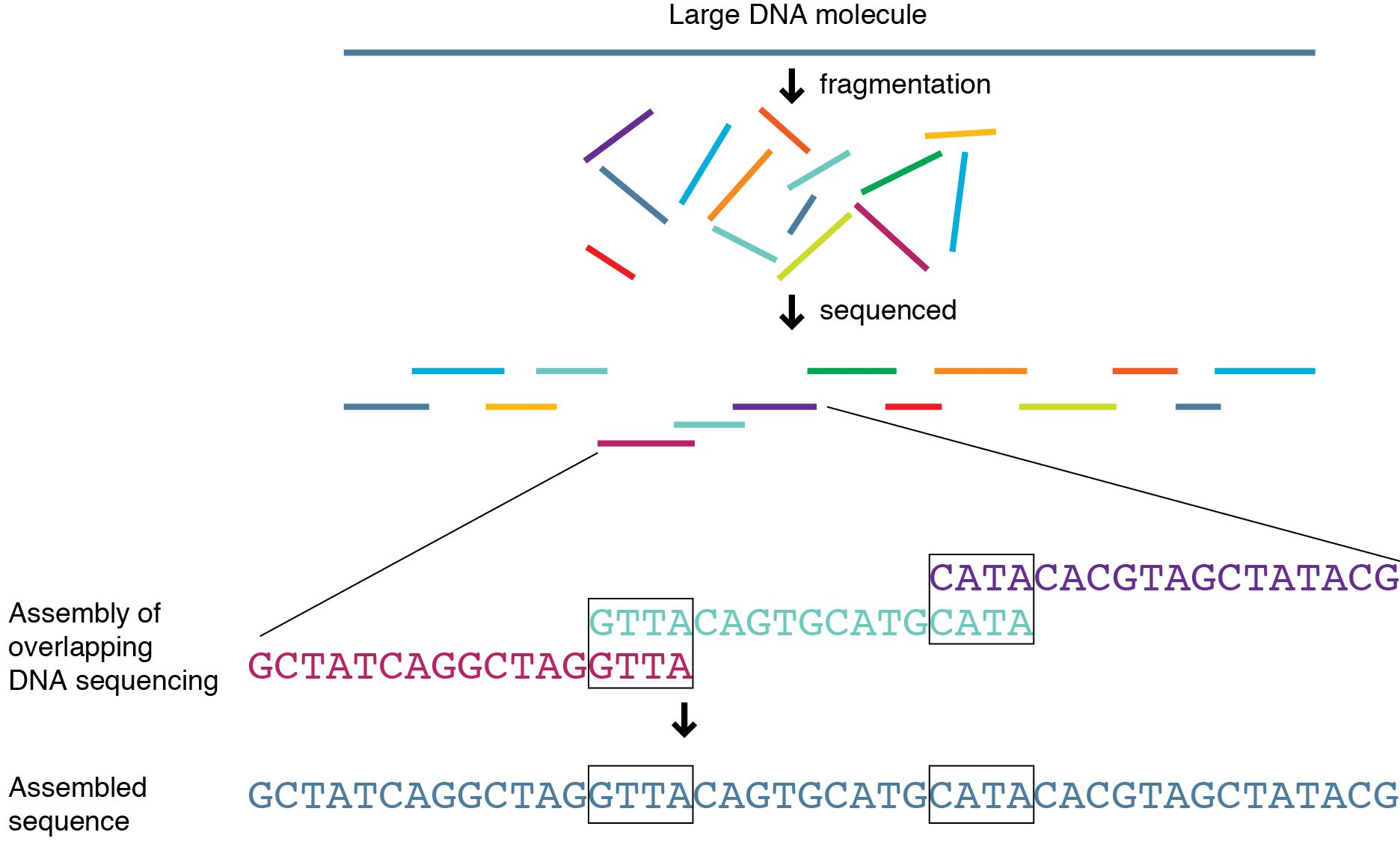
The complete sequence of a human genome sergey nurk 1,* , sergey koren 1,* , arang rhie 1,* , mikkorautiainen 1,* , andrey v. In reality, in order to sequence a whole human genome, you need to generate a bunch of short “reads” (~100 base pairs, depending on the platform) and then “align” them to the reference genome.
If you know the sequence of the bases in an organism, you have identified its unique dna fingerprint, or pattern.
Human whole genome sequencing. Whole genome sequencing (wgs) decodes all 6.4 billion dna base pairs in the human genome including the complete set of all 20,000 genes, mitochondrial dna, and the y chromosome. The human genome is written in dna, and while your exact genome is unique to you, about 99.9% of it is identical across all people. The complete sequence of a human genome sergey nurk 1,* , sergey koren 1,* , arang rhie 1,* , mikkorautiainen 1,* , andrey v.
Bzikadze 2 , alla mikheenko 3 , mitchell r. The 1000 genomes project, for instance, which on october 1 published details on the genomes of 2,504 individuals representing 26 populations, used both wgs and wes in its workflow, sequencing genomes to an average of 7x coverage and exomes to 66x [1]. What is whole genome sequencing (wgs)?
Whole genome sequencing, on the other hand, looks at all 3.2 billion base pairs (or 6.4 billion, depending on if you count both sets of chromosomes) in the human genome. Vollger 4 , nicolas altemose 5 , lev uralsky 6,7 ,ariel gershman 8 , sergey aganezov 9 , savannah j. And some researchers sequence genomes whole but use exon boundaries to constrain downstream.
If you know the sequence of the bases in an organism, you have identified its unique dna fingerprint, or pattern. We demonstrate that the data obtained is of high quality, with an average sequencing depth of 36.41, and that the output is comparable to data generated internationally on a similar platform. Here we report the whole genome sequences of the first six human dna samples sequenced and analysed at the south african medical research council�s genomics centre.
The genome, or genetic material, of an organism (bacteria, virus, potato, human) is made up of dna. In reality, in order to sequence a whole human genome, you need to generate a bunch of short “reads” (~100 base pairs, depending on the platform) and then “align” them to the reference genome. Human whole genome sequencing gain comprehensive access to human genetic variation pacbio highly accurate long reads let you generate complete and phased human genome assemblies of diverse populations to better understand the complexity of health and disease.
The sequencing of the first human genome cost over $3 billion,. As you might imagine, this provides a much bigger picture to. Each organism has a unique dna sequence which is composed of bases (a, t, c, and g).
It often helps obtain breakthrough achievements, which is applied more widely especially in the research of monogenic disease and cancer. Genomic information has been instrumental in identifying inherited disorders, characterizing the mutations that drive cancer progression, and tracking disease outbreaks. Since wgs is unquestionably becoming a new foundation for molecular analyses, we decided to compare three currently used tools for variant calling of human whole.
These findings show that chromothripsis is a major process that drives genome evolution in human cancer. Wgs has the ability to evaluate every base in the genome and navigate the complexity of genomic variants that make us unique. A genome is like a genetic instruction manual — it contains all the information an organism needs to grow and function.
Hawkins, in basic science methods for clinical researchers, 2017 overview of whole genome sequencing (wgs) wgs is the most global approach to identifying genetic variations. 1 the human genome project was the largest collaborative international biology project of its time.